RNAdecayCafe
Summary
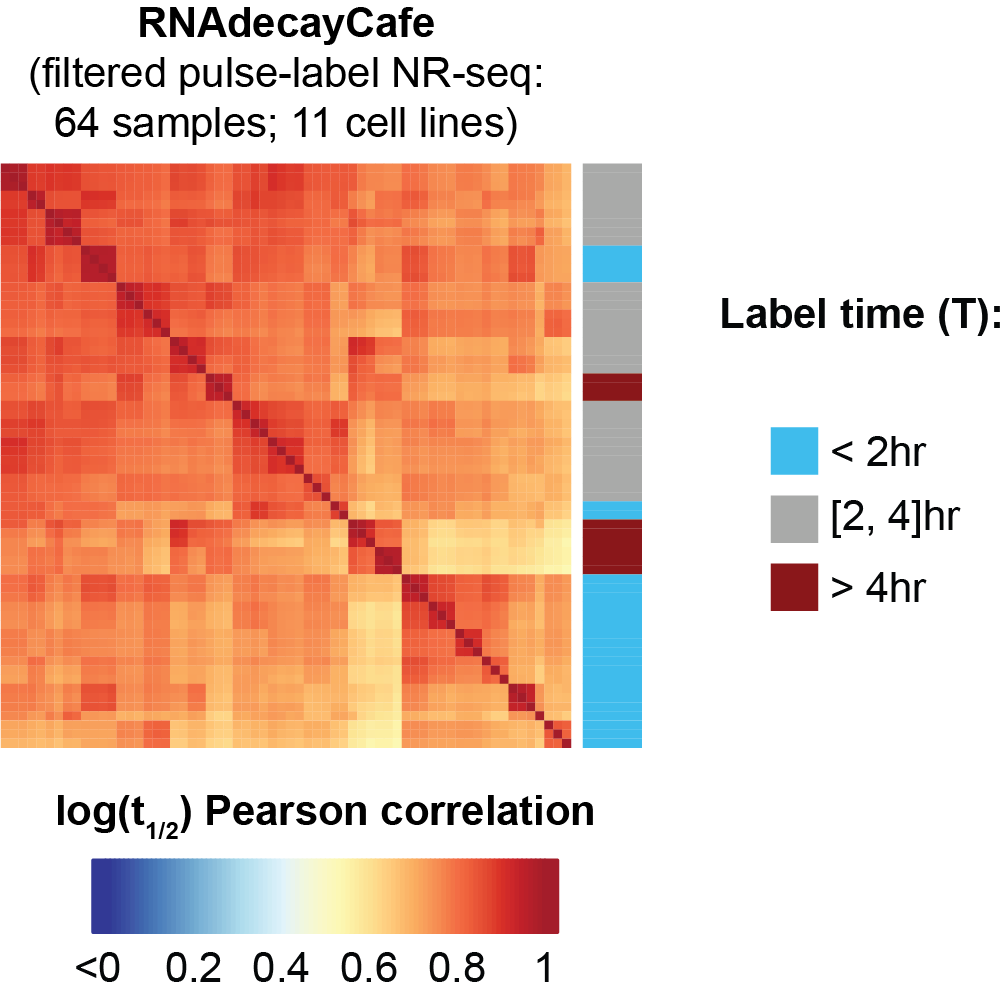
- Presents a first-of-its-kind database of high quality RNA stability estimates from pulse-label NR-seq experiments. Data can be accessed here.
- Paper presents a meta-analysis that shows how pulse-label NR-seq data is often highly consistent across labs, cell lines, method variations, etc. In contrast, we found pulse-chase NR-seq data to be batch effect-ridden and difficult to consistently interpret.
- We used this data to highlight the sometimes underappreciated role of RNA stability in tuning gene expression levels within a given cell line and in establishing differences across cell lines.
Link to preprint
The EZbakR-suite
Summary
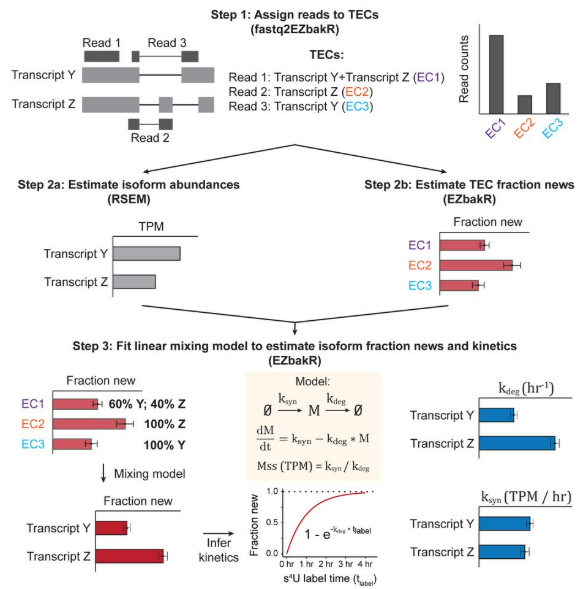
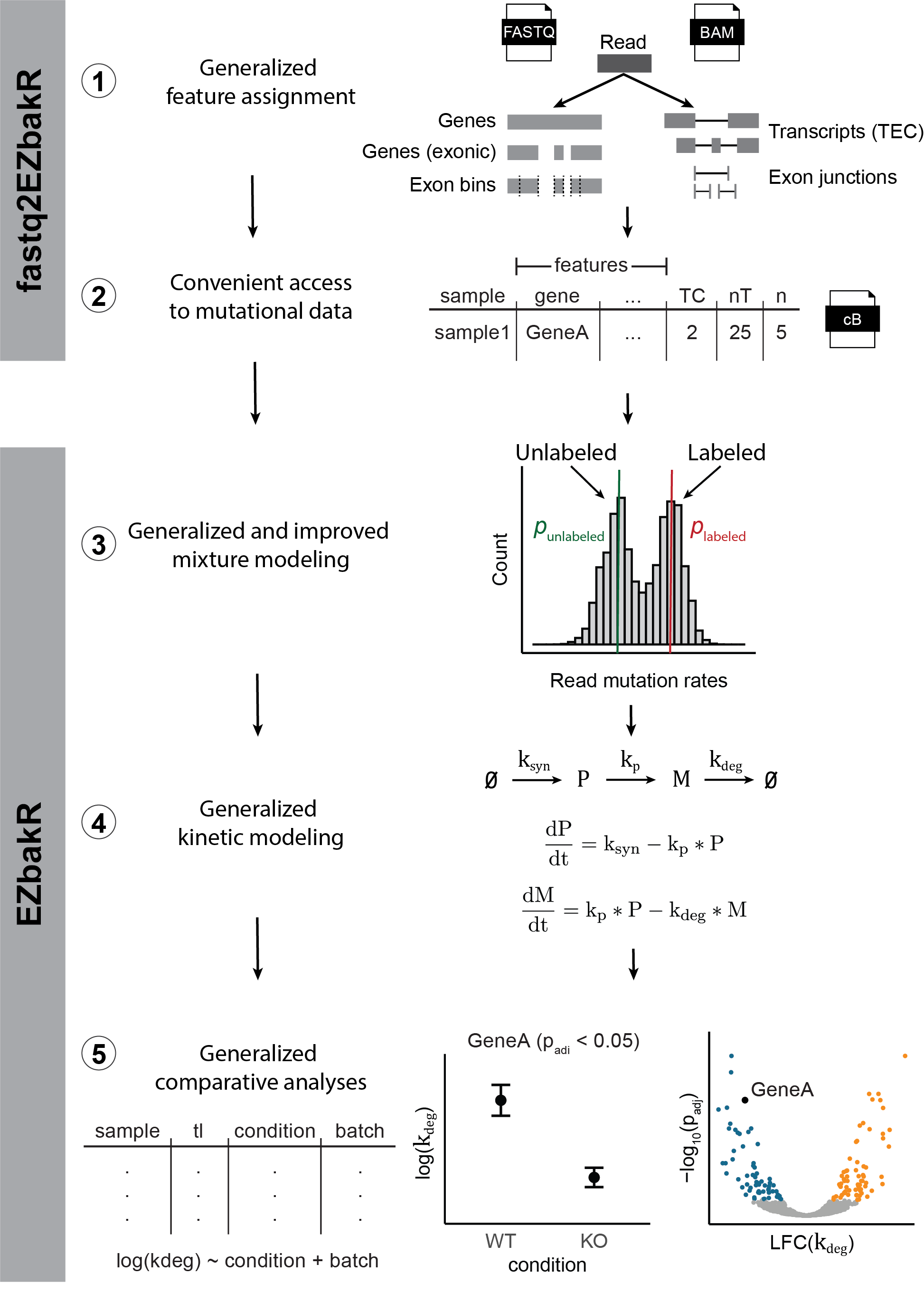
- Introduced Snakemake pipeline (fastq2EZbakR) and R package (EZbakR)
- Generalizes feature assignment: the set of annotated genomic features that you can perform analyses on.
- Generalizes mixture modeling: how the fraction of reads from labeled RNA are estimated.
- Generalizes dynamical systems modeling: how kinetic parameters are estimated from NR-seq data.
- Generalizes comparative analyses: how kinetic parameters are compared across biological conditions.
Link to paper
bakR
Summary
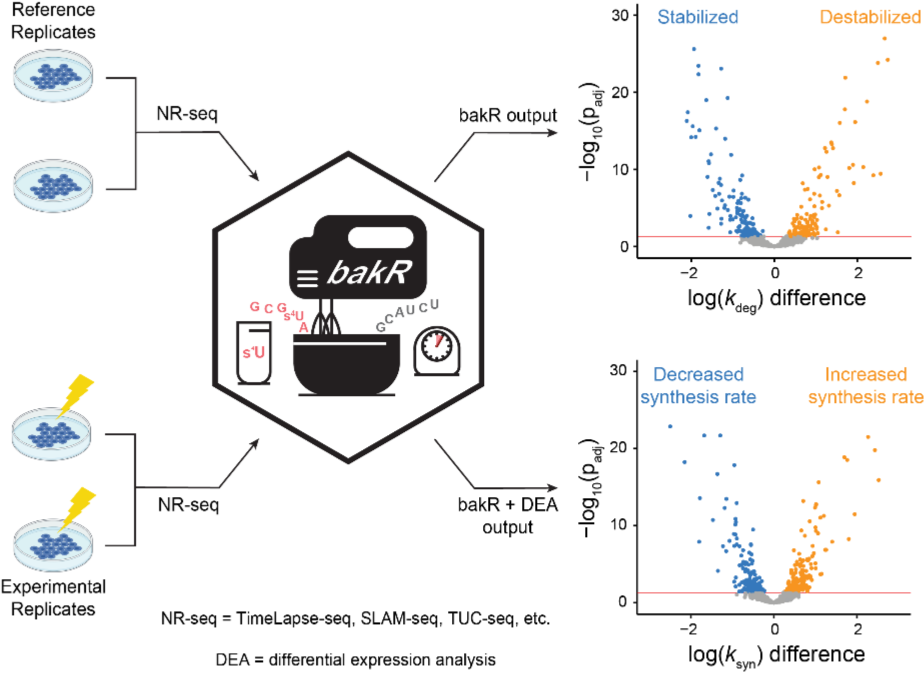
- Introduced an R package (bakR) designed to compare RNA synthesis and degradation rate constants across biological conditions.
- Also introduced a hierarchical modeling strategy to increase statistical power of these comparisons.
Link to paper